Key points
- CRISPR is an efficient, targetable gene editing tool which can be used to make genetic or epigenetic modifications, modulate gene expression or label gene loci.
- Cell and animal models created using CRISPR have provided insights into neurological disorders including autism, Parkinson’s disease and schizophrenia, and into neurological processes such as synapse formation.
- The use of CRISPR in a living brain involves challenges including delivery of CRISPR components, off-target effects and inefficient DNA repair machinery.
Abstract
Since it was first used to edit the mammalian genome,1 the targetable gene editing tool CRISPR (clustered regularly interspaced short palindromic repeats) has become widely accessible to researchers. Compared to older gene editing technologies, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), CRISPR has significant advantages: it is more efficient, faster to set up, and can be multiplexed: several DNA loci can be targeted in one experiment.2 CRISPR’s potential in neuroscience ranges from investigating fundamental processes underlying brain function and development, to modelling neurological diseases in both animals and cells, and perhaps to CRISPR-based therapies. This review discusses CRISPR’s current applications in cell and animal models aiming to clarify brain function and dysfunction, and some of the challenges that currently limit CRISPR’s use in neuroscience.
/sep
What is CRISPR?
The CRISPR gene editing system has been identified in and is derived from part of the prokaryote adaptive immune system, which defends against invading viruses or plasmids by specifically cleaving exogenous DNA. Adapting CRISPR for gene editing exploits the ability of CRISPR nucleases to make predictable DNA breaks at specifically targeted sequences. There are three types of CRISPR system (I, II and III) of which type II is most widely used in gene editing (see Figure 1).
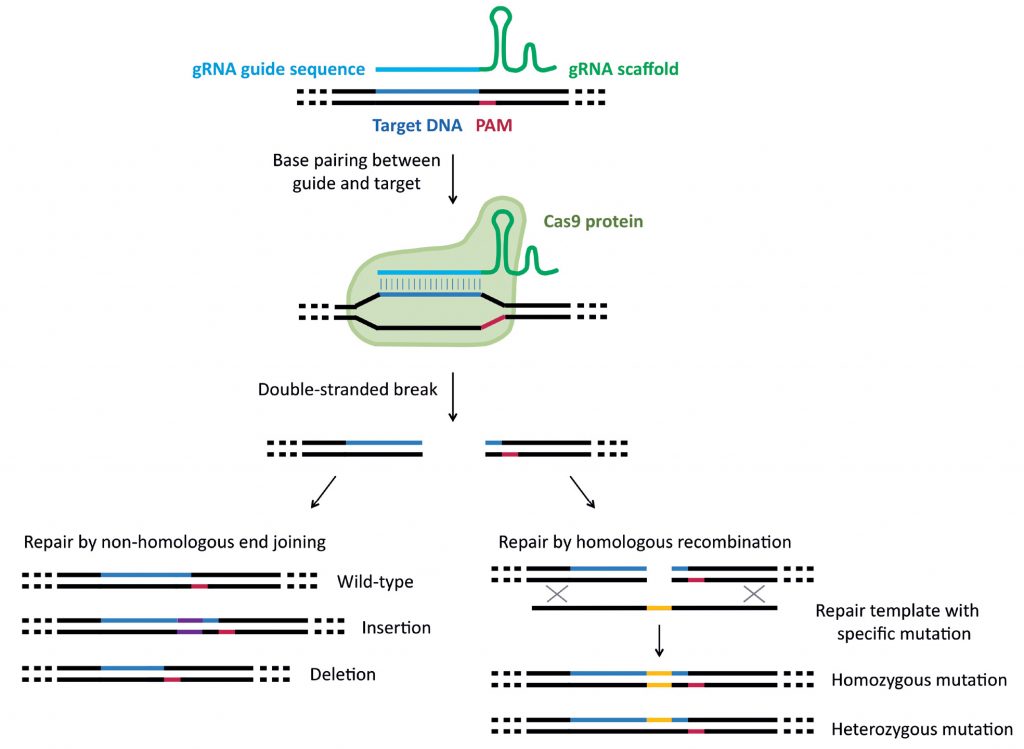
Guide RNA (gRNA) is composed of a scaffold sequence (required for binding between Cas9 and the gRNA) and a 20 base pair sequence which is designed complementary to the DNA target. The DNA target must be upstream of a proto-spacer adjacent motif (PAM) sequence, which is required for DNA cleavage. Cas9 nuclease is directed to the target DNA by base pairing between the target DNA and gRNA, resulting in a double-stranded DNA break. The double-stranded break can be repaired by non-homologous end-joining (NHEJ) or homologous recombination (HR). NHEJ results in either restoration of the wild-type sequence, insertions or deletions (indels). Indels vary in length and can cause frame-shift mutations, leading to premature stop codons and gene knockout. HR requires a DNA template with homologous regions up- and down-stream of the break. Therefore, a DNA repair template can be designed in order to introduce precise insertions, deletions or point mutation
First, guide RNA is designed complementary to a DNA target. Next, the guide RNA complexes with Cas9 nuclease (the CRISPR effector), followed by base-pairing between the guide RNA and its DNA target, which directs Cas9 to cleave the DNA. Finally, double stranded DNA breaks are repaired by either non-homologous end-joining (NHEJ) or homologous recombination (HR). NHEJ makes insertion or deletion mutations (indels) of varying lengths, usually resulting in a premature stop codon and gene knockout. Alternatively, DNA with the desired insertion, deletion or point mutation can be introduced to act as a repair template during HR, leading to precise mutations in the DNA.3
Cas9 is a nuclease, but CRISPR is not limited to nuclease activity. Inactivation of both catalytic domains in dead (d)Cas9 renders the nuclease unable to cleave DNA, but it can prevent transcription by steric hindrance in CRISPR interference (CRISPRi). CRISPRi can be enhanced by complexing dCas9 to repressors, for reversible gene knockdown, whereas dCas9-activator complexes can be used for reversible overexpression. In addition, dCas9 complexed to epigenetic modifiers can be used for methylation or histone modifications, and Cas9 with a fluorescent molecule can tag genomic loci.4 The range of functional domains coupled to Cas9 is expanding, linking CRISPR to advances in our understanding of genetic processes and our ability to manipulate them. One area where CRISPR has been readily adopted is in modelling neurological disease with human induced pluripotent stem cells (hiPSCs).
Cell models: hiPSCs
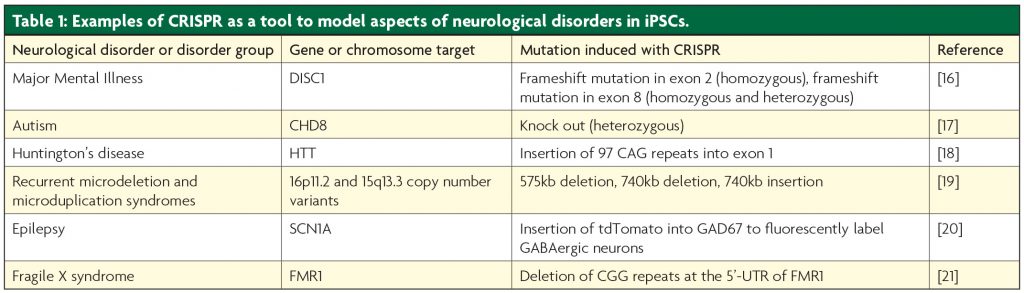
hiPSCs are somatic cells reprogrammed to an embryonic stem cell-like state, which retain the donor’s genetic identity and can make any cell type. hiPSCs from a donor with a neurological disease allow disease processes in cells usually inaccessible in a living patient to be studied in hiPSC-derived astrocytes, glia and neurons. CRISPR, ZFNs and TALENs can enhance hiPSC models by correcting or introducing genetic aberrations linked to a particular disorder, creating isogenic hiPSC lines, in which a specific genetic change can be studied without confounding genetic background effects. Isogenic hiPSC models have been made for multiple neurological disorders (see Table 1), including schizophrenia and Parkinson’s disease, 5 and offer a platform for drug screening, as well as for mapping pathways affected by disease-causing mutations.
In addition to DNA mutations, epigenetic changes, which alter gene expression without affecting the DNA sequence, have been implicated in neurological disorders including Alzheimer’s disease and epilepsy, as well as in neurological processes such as memory and cognitive aging.6 Several groups have made CRISPR-induced epigenetic modifications in cell models: for example, dCas9 fused to a histone demethylase has been targeted to gene enhancers, where it reduced gene expression in human cells.7 This proof of concept suggests that CRISPR could be used to study disease-linked epigenetic changes in cell models.
Animal models
Cell models are useful to study particular cell types in isolation, but animal models can give a more physiological representation of the human brain. The use of small animals, with relatively simple nervous systems, could be extended by using CRISPR to target multiple loci at once in a reverse genetic screen, in which indel mutations are made to identify genes with roles in a particular process. One group has used multiplexed guide RNAs to make indels at 48 loci thought to be involved in synapse formation in zebrafish, leading to the identification of two novel genes.8
CRISPR also facilitates the creation of larger animal models, which can otherwise be a lengthy and expensive process, particularly when multiple mutations are required. CRISPR has been used to make up to five mutations simultaneously in mouse embryonic stem cells, without apparent off-target effects.9 Genes can be targeted with CRISPR in vivo in existing mouse models, which can be aged to study aging-related neurological changes. Three genes involved in learning and memory have been simultaneously knocked out using CRISPR in a live adult mouse brain,10 showing that changes can be made in assembled neural circuits, which is an important step towards using CRISPR therapeutically in the brain.
Mouse models have provided valuable insights into neurological disorders, but their relevance to humans is limited by their relatively fast brain development, short lifespans and in some cases by gene expression under exogenous promotors. Larger mammalian models have brains closer in size and complexity to humans, and lifespans long enough to study aging-associated neurological diseases: pig and non-human primate models have been made to study Alzheimer’s, Huntington’s, and Parkinson’s diseases, amongst others.11 These models used one causative mutation, but CRISPR can be used to make multiple mutations in the same animal, allowing the study of multifactorial diseases or subtle phenotypes: in transgenic pigs, simultaneous mutations have been made in Parkin, DJ-1 and PINK1 (genes linked to early-onset Parkinson’s disease).12 In one cell monkey embryos indels have been made in two endogenous genes, which (although they are not linked to a particular human disorder) indicate the potential to alter multiple endogenous genes related to neurological diseases.13
Limits of CRISPR in the brain
CRISPR is a flexible and widely available tool which has been used to induce multiple specific disease-relevant mutations in cell and animal models of neurological disease. It has the potential to extend the use of current models through reversible modulation of gene expression and through epigenetic modifications. However, several challenges must be overcome for CRISPR to be used in a living brain. For instance, off-target effects are undesirable in animal models, and would be a safety concern if CRISPR was used therapeutically. Additionally, in vivo delivery of CRISPR components to the brain is difficult. Viral delivery is limited by the virus’s cloning capacity,10 but other methods, such as liposomal delivery of Cas9 protein and gRNA, have been used for gene knockout in the mouse inner ear in vivo.14 Finally, the correction of mutations or deletions by HR (following a CRISPR-induced DNA break) requires efficient DNA repair machinery, which may be less active in post-mitotic cells like neurons.15 CRISPR-mediated changes which do not require DNA repair, such as epigenetic modifications or CRISPRi, may therefore be more easily induced in neurons in vivo. Alternatively, NHEJ, which does occur in neurons, could be used to make indels for therapeutic gene knockout in disorders caused by toxic gain of function, such as Huntington’s disease.
Some of these problems may be overcome by utilising alternative CRISPR systems as gene editing tools. As more prokaryote genomes are sequenced, CRISPR or CRISPR-like systems with different effectors or DNA cleavage characteristics may come to light, which could further diversify CRISPR’s gene editing potential. Even without new CRISPR systems, the growing family of CRISPR-based tools clearly indicates that CRISPR is yet to reach its full potential in neuroscience.
References
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–23. doi:10.1126/science.1231143.
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347–55. doi:10.1038/nbt.2842.
- 3. Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–308. doi:10.1038/nprot.2013.143.
- Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 2015;17:5–15. doi:10.1038/nrm.2015.2.
- Russo FB, Cugola FR, Fernandes IR, et al. Induced pluripotent stem cells for modeling neurological disorders. World J Trasnplant 2015;5:209–21. doi:10.1002/biot.201400010
- Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–32. doi:10.1016/j.neuron.2013.10.023.
- Vojta A, Dobrinić P, Tadić V, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 2016;:1–14. doi:10.1093/nar/gkw159.
- Shah AN, Davey CF, Whitebirch AC, et al. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods 2015;12:535–40. doi:10.1038/nmeth.3360.
- Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/cas-mediated genome engineering. Cell 2013;153:910–8. doi:10.1016/j.cell.2013.04.025.
- Swiech L, Heidenreich M, Banerjee A, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 2015;33:102–6. doi:10.1038/nbt.3055.
- Tu Z, Yang W, Yan S, et al. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener 2015;10:35. doi:10.1186/s13024-015-0031-x.
- Wang X, Cao C, Huang J, et al. One-step generation of triple gene- targeted pigs using CRISPR/Cas9 system. Nat Publ Gr 2016;:1–7. doi:10.1038/srep20620.
- Niu Y, Shen B, Cui Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014;156:836–43. doi:10.1016/j.cell.2014.01.027.
- Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 2014;33:73–80. doi:10.1038/nbt.3081.
- McKinnon PJ. DNA repair deficiency and neurological disease. Nat Rev Neurosci 2009;10:100–12. doi:10.1038/nrn2559.
- Srikanth P, Han K, Callahan DG, et al. Genomic DISC1 Disruption in hiPSCs Alters Wnt Signaling and Neural Cell Fate. Cell Rep 2015;12:1414–29. doi:10.1016/j.celrep.2015.07.061.
- Wang P, Lin M, Pedrosa E, et al. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in neurodevelopment. Mol Autism 2015;6:55. doi:10.1186/s13229-015-0048-6
- An MC, O’Brien RN, Zhang N, et al. Polyglutamine Disease Modeling: Epitope Based Screen for Homologous Recombination using CRISPR/Cas9 System. PLoS Curr 2014;6. doi:10.1371/currents.hd.0242d2e7ad72225efa72f6964589369a.
- Tai DJC, Ragavendran A, Manavalan P, et al. Engineering microdeletions and microduplications by targeting segmental duplications with CRISPR. Nat Neurosci 2016;19:517–22. doi:10.1038/nn.4235.
- Liu J, Gao C, Chen W, et al. CRISPR/Cas9 facilitates investigation of neural circuit disease using human iPSCs: mechanism of epilepsy caused by an SCN1A loss-of-function mutation. Transl Psychiatry 2016.
- Park CY, Halevy T, Lee DR, et al. Reversion of FMR1 Methylation and Silencing by Editing the Triplet Repeats in Fragile X iPSC-Derived Neurons. Cell Rep 2015;13:234–41. doi:10.1016/j.celrep.2015.08.084.
