Summary
- Data-driven models provide a uniquely fine-grained multi-modal picture of disease progression.
- This offers major potential benefits to neurodegenerative disease research and clinical practice, by improving patient staging and monitoring, disease prognosis and differential diagnosis.
- To date these models have provided valuable insights into neurodegenerative disease progression patterns, particularly in Alzheimer’s disease.
- Data-driven models are an emerging area of technology with numerous exciting opportunities for future developments.
- These techniques have wide potential further application to any disease or developmental process.
Neurodegenerative diseases are characterised by the temporal order and severity of a distinct set of symptoms and pathological changes that occur within the brain. Whilst the underlying mechanisms by which these pathologies arise and propagate are not fully understood, the development of imaging and CSF measures that reflect the presence and severity of these pathological changes is providing valuable insights, including opening a potential pre-symptomatic window where disease-modifying therapies may be most effective. Characterising the trajectories of these biomarkers over the time course of different neurodegenerative diseases is of great interest in order to build up a quantitative picture of disease progression.1 Such a picture provides insight into the underlying disease biology and, moreover, provides a potential mechanism for patient staging and monitoring, disease prognosis and differential diagnosis.
Recently a range of hypothetical models have been proposed that describe the long-term progression of biomarkers associated with different neurodegenerative diseases, with a particular focus on Alzheimer’s disease1-3 (Figure 1). However, a fully quantitative data-driven model is required for practical application to patient staging and monitoring, prognosis and differential diagnosis. Wide recognition of the need for diverse multi-biomarker data sets to inform such quantitative progression models has lead to the establishment of large multi-centre biomarker studies including ADNI (sporadic Alzheimer’s disease),4 DIAN (familial Alzheimer’s disease),5 predict-HD (Huntington’s disease),6 PPMI (Parkinson’s disease),7 and many others. However, reconstructing biomarker trajectories from these data sets is challenging. The data are largely cross-sectional or with only a few years of follow up available, which is a short time period relative to the long disease time course that may span several decades. Reconstructing biomarker trajectories from these data sets requires new modelling techniques that can bring together cross-sectional and short-term longitudinal data at unknown time points to reconstruct a common progression pattern across subjects. Further challenges arise from misdiagnosis (either cases not having the disease in question, or controls having pre-symptomatic disease), mixed pathology, and sparsity of data points at the beginning and end of the disease time course.
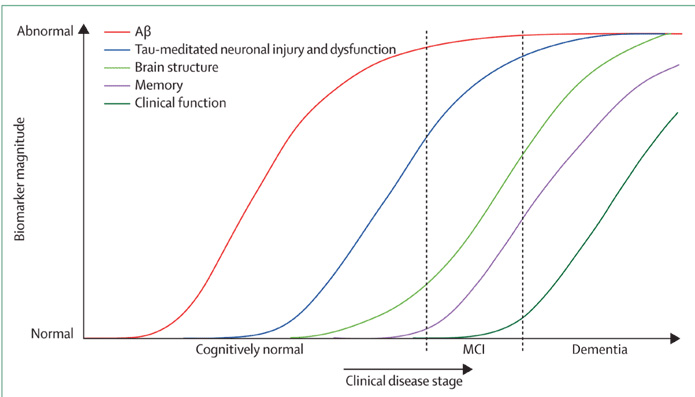
Traditional statistical analysis techniques estimate biomarker trajectories by assuming a priori knowledge of where each data point lies along the disease time course. Hence, the majority of studies of neurodegenerative disease biomarker progression8,9 rely on the use of a priori clinical classification as a patient staging measure and then compare biomarkers across groups. This reliance on clinical staging limits the temporal resolution of the biomarker progression to only a few stages, e.g. in Alzheimer’s disease: ‘cognitively normal’, ‘mild cognitive impairment’ and ‘Alzheimer’s disease’. Recently a new family of truly data-driven statistical models10-12 have emerged that do not require prior knowledge of the stage of each individual along the disease time course. This is a major advantage, as it allows for a complete picture of disease progression incorporating the full set of biomarkers, and with much higher temporal resolution. In this review we focus on these models, giving an overview of the different types that have been applied to neurodegenerative diseases so far; and the future potential of such data-driven disease progression models.
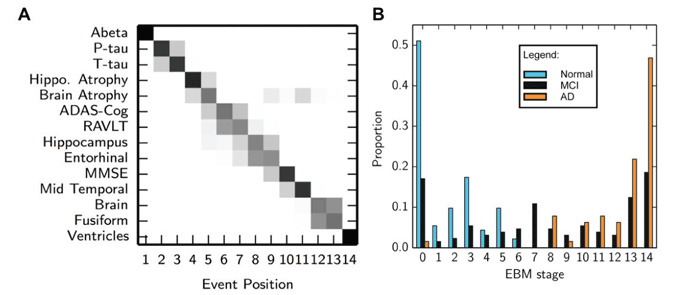
The event-based model10 describes disease progression as a series of events, where each event corresponds to a particular biomarker becoming abnormal. The unique property of the event-based model is that it directly encodes, and thus estimates from the data, the ordering in which biomarkers become abnormal, or, more strictly, observably different from normal levels. This sequence of events provides a simple and intuitive description of disease progression, as well as a natural patient staging system – at stage X, the first X events have occurred. The event-based model has been applied to recover the sequence of regional neurodegeneration in both familial Alzheimer’s disease and Huntington’s disease.10 More recently it has been modified for the more challenging application to sporadic neurodegenerative diseases13 (Figure 2), and applied to determine the sequence of abnormality in sporadic Alzheimer’s disease for a multi-modal set of biomarkers, including CSF measures of amyloid-beta and tau, regional volumetric and rates of atrophy measures from MRI, and cognitive test scores. Young et al13 further demonstrate the clinical utility of the event-based model as a patient staging system, providing state-of-the-art classification accuracy for separating cognitively normal and Alzheimer’s disease subjects, and for predicting conversion from cognitively normal to mild cognitive impairment and mild cognitive impairment to Alzheimer’s disease. Another key strength of the event-based model is its probabilistic formulation, which provides measures of confidence in both the sequence of biomarker abnormality events across the population, and an individual’s model stage. The event-based model naturally extends to differential diagnosis by providing a likelihood of each candidate neurodegenerative disease, which is achieved by fitting an individual’s set of biomarker measurements to each corresponding biomarker sequence. One limitation of the event-based model is that it doesn’t incorporate any information on the time between events or the rate of biomarker decline, which somewhat limits its utility for prognosis and monitoring.
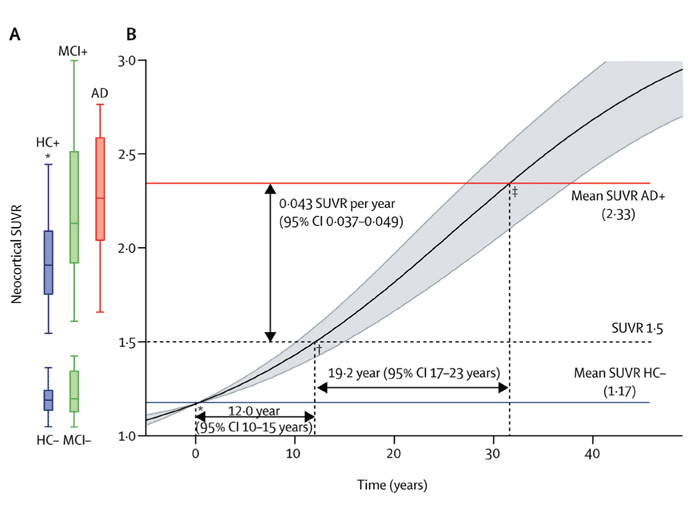
Differential equation models11,14-17 can be used to reconstruct an average cohort-level biomarker trajectory, which is continuous in contrast to the discrete description of the event-based model. The models use short-term follow up biomarker measurements to provide samples of the gradient of a single common biomarker trajectory and integrate a differential equation to determine a best-fit or ‘average’ trajectory for the cohort. For example, Jack et al17 determine the time taken for amyloid accumulation to go from a normal to an abnormal level by fitting a differential equation model to data from serial amyloid-PET scans, finding that it takes approximately 15 years to go from a normal standard uptake value ratio (SUVR) of 1.5 to an abnormal SUVR of 2.5. Villemagne et al11 (Figure 3) perform a similar analysis to determine the time taken for several biomarkers to go from normal to abnormal, including amyloid-PET, hippocampal atrophy, episodic memory, gray matter volume and non-memory cognitive domains. Differential equation models have potential as a disease staging, monitoring and prognostic tool as they provide the rate of biomarker decline over the disease time course. Stochastic differential equation models18 can further express deviations from this average, providing prognostic information at the individual level. However, they model each biomarker individually, and so there is no guarantee of correspondence across disease stage and prognosis estimates between different biomarkers.
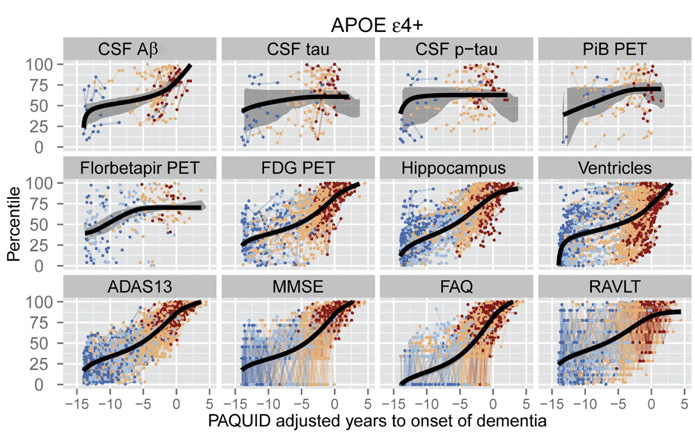
Self-modelling regression approaches12,19 bring together data from multiple biomarkers to estimate biomarker trajectories over a common disease timescale. Short-term follow up data from each individual provides samples of a common set of biomarker curves, which are used to estimate the population-level shape and rate of biomarker decline, as well as each individual’s position and rate of decline. As with differential equation models, the biomarker curves represent the average biomarker dynamics for a population. Donohue et al12 (Figure 4) use self-modelling regression to determine the trajectories of cognitive test scores, regional brain volumes from MRI, PET imaging measures, and CSF levels of amyloid-beta and tau. Jedynak et al19 formulate a similar model that uses cognitive test scores, CSF amyloid-beta and tau, and hippocampal volume on MRI to estimate a ‘disease progression score’, which is a continuous measure of disease stage that can be used as a time proxy. Self-modelling regression approaches provide continuous disease staging, monitoring and prognostic measures that incorporate information from multiple biomarkers. A key advantage of these models is that they provide a very complete picture of the disease, which can aid detailed disease understanding. Potential disadvantages are that they have many more parameters to estimate than simpler models like the event-based model, so may be less stable; and the complex picture has a less straightforward interpretation than the discrete description, which may limit clinical utility.
To date, these data-driven models have shown compelling results that provide valuable insights into neurodegenerative disease progression patterns, particularly in Alzheimer’s disease. However, they remain an emerging area of research, and all the current models share a number of limitations and assumptions that are important to consider when interpreting results. One strong assumption that all the aforementioned models make is that all subjects follow a common progression pattern. Although some models allow for subjects to deviate from this common progression pattern, these deviations are assumed to be small, and none allow for subgroups of subjects that follow completely different progression patterns. Such outliers are likely given the inherent heterogeneity of sporadic disease data sets, which contain some proportion of subjects with alternative neurodegenerative diseases, as well as mixed pathologies and a wide range of subject demographics. For this reason, practical applications of data-driven models often focus on more homogeneous population subgroups,11-13,17 for example subjects with increased genetic risk of developing the neurodegenerative disease of interest. Another assumption is the independence of biomarkers: although the models express temporal correlation of biomarker trajectories over the disease time course, they assume independence at any given time point. In practice, biomarkers often co-vary, for example amyloid-PET and CSF measures of amyloid-beta are measures of the same underlying pathology and are therefore strongly correlated. Failure to model this covariance tends to cause underestimation of the variance of progression patterns across the population. Data-driven models further assume that data is available from the full disease time course when in reality the data points may be sparse at the beginning and end of the disease progression, which may influence the estimation of biomarker trajectories.
Future developments in disease progression modelling offer numerous exciting opportunities. Adaptations to characterise the heterogeneity in sporadic disease data sets are certainly possible, for example by using mixture models or distributions of event-sequences or biomarker trajectories, which is desirable for the application of these models on an individual level in clinic. This will help separate measurement noise from inter- and intra-subject variation depending on genetic, lifestyle and demographic information. The high temporal resolution of data-driven models is promising for their use in patient staging and disease monitoring. The discrete stages of models like the event-based model align well with general medical practice, but continuous models provide more useful prognostic information. Future models designed for clinical use might combine elements of both, allowing continuous prognostic estimates, but also subdividing the progression into discrete stages. Data-driven models also present exciting opportunities for differential diagnosis; the ideas are somewhat robust to common problems such as missing data and differing study designs, so the models provide a natural framework for making information about different neurodegenerative diseases compatible. Such work will also enhance basic disease understanding by highlighting the most discriminative features for differential diagnosis. New types of model yet to be explored include spatiotemporal models (e.g. network models20), which so far have relied on a priori clinical staging, but new approaches are emerging that may help avoid this limitation.21,22
Data-driven models are an emerging area of technology with major potential benefits to neurodegenerative disease research and clinical practice, and with wide potential further application to any disease or developmental process. They can provide quantitative multi-modal pictures of the full disease time course for improved understanding of disease mechanisms to inform drug discovery; they naturally combine different types of information for earlier and more accurate differential diagnosis, and subject-specific prognostic information; they provide fine-grained staging scores or systems for more precise patient stratification supporting clinical trials for developing treatments and ultimately treatment deployment. Research is ongoing to refine this emerging technology into a practical tool in medical development and practice.
References
- Jack CR, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, Petersen RC, Trojanowski JQ. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 2010;9(1):119-28.
- Frisoni GB, Blennow K. Biomarkers for Alzheimer’s: the sequel of an original model. Lancet Neurol 2013;12(2):126-8.
- Aisen PS, Petersen RC, Donohue MC, Gamst A, Raman R, Thomas RG, Walter S, Trojanowski JQ, Shaw LM, Beckett LA, Jack CR, Jagust W, Toga AW, Saykin AJ, Morris JC, Green RC, Weiner MW. Clinical Core of the Alzheimer’s Disease Neuroimaging Initiative: progress and plans. Alzheimers Dement 2010;6(3):239-46.
- http://adni.loni.usc.edu/
- http://www.dian-info.org/
- https://www.predict-hd.net/
- http://www.ppmi-info.org/
- Thompson PM, Hayashi KM, De Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of Gray Matter Loss in Alzheimer’s Disease. J Neurosci 2003;23(3):994-1005.
- Scahill RI, Schott JM, Stevens JM, Rossor MN, Fox NC. Mapping the evolution of regional atrophy in Alzheimer’s disease: Unbiased analysis of fluid-registered serial MRI. Proc Natl Acad Sci;2002;99(7):1-5.
- Fonteijn HM, Modat M, Clarkson MJ, Barnes J, Lehmann M, Hobbs NZ, Scahill RI, Tabrizi SJ, Ourselin S, Fox NC, Alexander DC. An event-based model for disease progression and its application in familial Alzheimer’s disease and Huntington’s disease. Neuroimage 2012;60(3):1880-9.
- Villemagne VL, Burnham S, Bourgeat P, Brown B, Ellis KA, Salvado O, Szoeke C, Macaulay SL, Martins R, Maruff P, Ames D, Rowe CC, Masters CL. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: a prospective cohort study. Lancet Neurol 2013;12(4):357-67.
- Donohue MC, Jacqmin-Gadda H, Le Goff M, Thomas RG, Raman R, Gamst AC, Beckett LA, Jack CR, Weiner MW, Dartigues J-F, Aisen PS. Estimating long-term multivariate progression from short-term data.” Alzheimers. Dement 2014;1-11.
- Young AL, Oxtoby NP, Daga P, Cash DM, Fox NC, Ourselin S, Schott JM, Alexander DC. A data-driven model of biomarker changes in sporadic Alzheimer’s disease. Brain 2014;137:2564-77.
- Ashford JW, Schmitt FA. Modeling the time-course of Alzheimer dementia. Curr Psychiatry Rep;2001;3(1):20-8.
- Yang E, Farnum M, Lobanov V, Schultz T, Verbeeck R, Raghavan N, Samtani MN, Novak G, Narayan V, DiBernardo A. Quantifying the pathophysiological timeline of Alzheimer’s disease. J Alzheimers Di 2011;26(4):745-53.
- Sabuncu MR, Desikan RS, Sepulcre J, Yeo BTT, Liu H, Schmansky NJ, Reuter M, Weiner MW, Buckner RL, Sperling RA, Fischl B. The dynamics of cortical and hippocampal atrophy in Alzheimer disease. Arch Neurol 2011;68(8):1040-8.
- Jack CR, Wiste HJ, Lesnick TG, Weigand SD, Knopman DS, Vemuri P, Pankratz VS, Senjem ML, Gunter JL, Mielke MM, Lowe VJ, Boeve BF, Petersen RC. Brain β-amyloid load approaches a plateau. Neurology 2013;80(10):890-6.
- Oxtoby NP, Young AL, Fox NC, Daga P, Cash DM, Ourselin S, Schott JM, Alexander DC. Learning imaging biomarker trajectories from noisy Alzheimer’s disease data using a Bayesian multilevel model. Lecture Notes in Computer Science 2014;8677:85-94.
- Jedynak BM, Lang A, Liu B, Katz E, Zhang Y, Wyman BT, Raunig D, Jedynak CP, Caffo B, Prince JL. A computational neurodegenerative disease progression score: method and results with the Alzheimer’s disease Neuroimaging Initiative cohort. Neuroimage 2012;63(3):1478-86.
- Xie T, He Y. Mapping the Alzheimer’s brain with connectomics. Front. psychiatry 2011;2(1):77.
- Aljabar P, Wolz R, Srinivasan L, Counsell S, Boardman JP, Murgasova M, Doria V, Rutherford MA, Edwards AD, Hajnal JV, Rueckert D. A Manifold Learning Framework. Application to Neonatal MRI. 2010;1-8.
- Davis BC, Fletcher PT, Bullitt E, Joshi S. Population Shape Regression from Random Design Data. Int J Comput Vis 2010;90(2)255-66.



